We recently asked three experts what they thought about the use of artificial intelligence and machine learning in drug discovery, a topic that has been front and center lately, discussed regularly at scientific meetings as well as in mainstream publications. In the Q&A below, Grant Wishart, Ph.D., Director CADD & Structural Biology at Charles River Laboratories; Stephan Steigele, Ph.D., Head of Science Genedata; and Matt Segall, Ph.D., CEO at Optibrium, share their considerable insights and thoughts on the future.
Biocompare: Is the use of AI (as well as ML and DL) currently improving the drug discovery process?
Grant Wishart, Ph.D.: The application of AI technologies within drug discovery has great potential. The ability to interrogate large datasets, identify underlying patterns, make predictions, and convert to knowledge, ultimately evolving how we discover new drugs is very exciting. Currently the lines are blurred between hype and real potential. It will only be when such technologies are demonstrated in real-life drug discovery paradigms that the reality and hype will separate and the true value will emerge. Perhaps it is still too early to judge the real impact of AI in the drug discovery process.
Stephan Steigele, Ph.D.: Currently, powerful AI uses exist along the drug discovery process, and I have no doubt that many more applications are on the horizon. I find this to be a natural development and logical progression for AI in drug discovery. Life science and biopharma data, in particular, are best suited for AI and machine learning as there is an enormous amount of data that is structured and frequently well annotated. This high degree of available training data is the enabler for successful AI applications.
A lot of exciting and successful AI applications in drug discovery are making their mark on the industry. Recursion Pharmaceuticals is using AI to unlock the maximum amount of information, often deduced from cellular images, to dramatically improve and accelerate drug research and drug repurposing. This is resulting in an impressive number of candidates pushed into clinical trials. Another outstanding example of AI in drug discovery is from Janssen Pharmaceutica and their collaboration partners as published in Cell Chemical Biology. Here, they utilized massive high-content screening image data to predict compound effects on so far untested cell types and how they react to already tested chemical compounds, which gives Janssen a huge leg up when starting a new screening project.
Of course, my own experience with Genedata Imagence® demonstrates how AI-driven solutions can accelerate and streamline phenotypic assay analysis. So, these are some prime examples of how the promise of AI in drug discovery can be fulfilled right from the start when applied in an appropriate context.
Matt Segall, Ph.D.: There are several promising applications of AI to drug discovery, but so far I think that the impact has been limited. One problem has been that many scientists have tried to apply off-the-shelf ML/DL methods to drug discovery data and found that the improvements over conventional methods have been small. This is because the data in drug discovery are very different from those in the domains in which the ML/DL methods were initially developed. The data available in drug discovery projects are much more sparse and noisy than in, say, image or voice recognition.
Probably the greatest impact so far has come from text and natural language processing to enable more powerful search for relevant data in the literature or patents. This has greater parallels with applications in other domains than application of ML/DL to predictive modelling.
Biocompare: At this time, what is the most advanced or valuable application of AI (ML/DL) in drug discovery?
Grant Wishart, Ph.D.: AI methodologies to support drug discovery are emerging at a rapid pace, but lag behind other industries with respect to application and maturity. At Charles River, technologies in the early discovery space are being evaluated to ensure only those with the highest potential are adopted for our partners. This is reflected in our recently announced strategic partnership with Atomwise, providing Atomwise’s unique structure-based AI technologies for hit identification and optimization to our partner projects.
Stephan Steigele, Ph.D.: I believe the most spectacular report during the past year was around Google DeepMinds AlphaFold, which simply elevated the quality of de-novo protein structure predictions (by just starting with a DNA/Protein sequence as input) beyond any previous level. What I really found spectacular is that this work wasn’t delivered by domain experts, such as structural biologists, bio-physicians, or chemists, but by a very small team of computer scientists. For me, this shows the power of the underlying approach. When I studied for my Ph.D. in computer science, the in-silico protein folding prediction problem was mentioned in every textbook as the holy grail of bioinformatics. I view Google’s team of computer scientists cleverly applying AI to come much closer to the holy grail than anyone else so far.
Many more valuable applications are coming that will close the loop between early discovery programs and late-stage activities. For example, helping pathologists with tumor diagnoses while closely monitoring successes and failures of drug candidate formulas during clinical trials (e.g., in lung cancer screening). Utilizing weak data connections, which are now observed as there are far more measurables than ever before, will contribute to improved control, mitigate investment risks, and strengthen personalized medicine.
Matt Segall, Ph.D.: There are a number of applications of AI that have recently shown significant advances and have the potential to offer great value in drug discovery:
1. We recently demonstrated a novel DL method for imputation of compound bioactivities and other properties that achieved significant improvements in accuracy over conventional quantitative structure-activity relationship models. In addition, this method can estimate the confidence in each prediction, enabling only the most confident predictions to be used to guide better decisions.
2. Recent advances in application of DL to chemical reactions are showing promise for the prediction of synthetic pathways. This will help in the assessment of synthetic tractability of new compounds and synthesis planning (see for example Schwaller et al., and Coley et al.)
3. The combination of de novo design with predictive models of compound properties and synthetic planning can help to guide a rigorous and objective exploration of compound-optimization strategies in the context of a drug discovery project (see for example Segall et al.)
Biocompare: Do researchers need to be concerned that AI (ML/DL) is going to make their jobs obsolete? What kind of training should scientists have to prepare for AI implementation in their labs?
Grant Wishart, Ph.D.: Researchers have a great opportunity to drive the process of increased automation and AI within their drug discovery environments. Using their knowledge of prior processes and past pitfalls from target discovery to drug approval, they can help identify areas where predictive technologies can make sense of vast data repositories allowing better decision making at speed. It is unlikely that this will render drug discovery research positions obsolete in the near future, however organizations will look to develop their staff internally and acquire an increasing number of individuals in areas such as data science, machine learning, and automation engineering. Within Charles River for example, we continuously train and develop current staff through extensive training programs and secondments. This is supplemented with recruitment of new staff with the skills we need. Thus allowing us to support individual personal development and fulfill ongoing business needs.
Stephan Steigele, Ph.D.: Speaking from experience as part of the development team of Genedata Imagence, this AI-based solution puts scientists in full control of the procedure while making them very efficient. Here was our development rationale: many research tasks don’t require human interaction. These tasks are tedious and repetitive and yet have required specific expertise, which results in a very time-consuming and error-prone process. We wanted to replace tedious and time-consuming steps with a revolutionary application of our AI. The interesting point with our solution: it enables scientists to find more relevant pharmacological endpoints than with any non-AI approach and in a shorter period of time. Our AI solution empowers scientists to use their time and energy for better science vs. low-level image analysis—the underlying process is handled by AI.
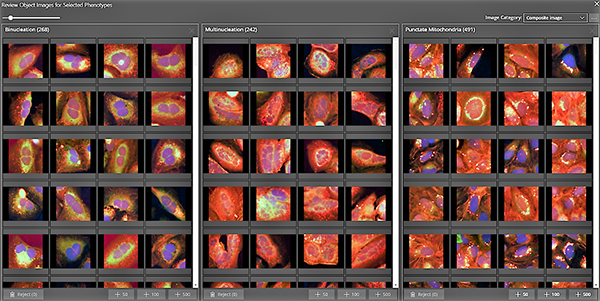
Image: An example of an AI application in early drug discovery (screenshot from Genedata Imagence). High-content screening showing three phenotype classes (binucleated, multinucleated, and punctate mitochondria)—found, defined, and enriched with deep learning-based technology (image courtesy of Genedata).
AI will change lab work for scientists. Integrating with existing workflows, AI will assist scientists in decision making rather than making the decision. Therefore, scientists need to learn to interpret AI predictions and understand the connection between prediction results and the underlying training data. They will need to distinguish meaningful AI predictions from predictions that simply are wrong. I’m confident that AI systems will become more robust in further assisting researchers. Our work and the work of many others show that AI systems by themselves are capable of judging the ‘quality’ of their own predictions; and therefore, can react and instruct humans by requesting more relevant training data with the aim of increasing the predictive relevance of results.
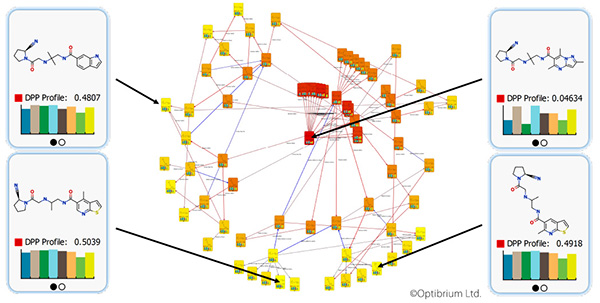
Matt Segall, Ph.D.: I do not think that AI will make researchers’ jobs obsolete. In the foreseeable future, the principle of augmentation will be much more important; it has been demonstrated in many disciplines that the combination of a human expert with an AI performs better than either individually. The impact of this will be that AI will help researchers to become more effective at their jobs. It will also reduce the amount of routine, repetitive work and free researchers to tackle the more scientifically challenging and interesting questions. The skills and experience of researchers will remain an important part of the process.
Image: Computational methods can augment scientists’ experience and help to explore a broad range of new compound optimization strategies. Image courtesy of Optibrium.
Biocompare: Is access to quality data an issue impeding adoption of AI (ML/DL)?
Grant Wishart, Ph.D.: It is generally recognized that the time spent collating and preparing data far exceeds the data analysis, model building, and validation processes. As a result, access to high-quality, curated, and preferably homogeneous data is always a priority. At Charles River, standardization of data storage and processing is being implemented to facilitate optimal data use. As a services company, data is most often owned by our client partners who we are working alongside to leverage through model exploration and analysis.
Stephan Steigele, Ph.D.: Access to quality data is indeed the most striking constraint for the successful development and application of AI-driven solutions. This is the main reason that pharma companies are highly engaged in AI-driven research as most of their data are easily accessed by internal teams only.
To be clear, I believe the lack of public training data is a problem: without such data, algorithms and their approaches are understood with a limited scope, especially when their reproducibility and required refinements are not achievable due to a lack of data. Independent confirmation of findings is commonly agreed upon as the hallmark of modern science. I think that both academic and commercial parties must start thinking about high-quality data including high-quality labels shared during pre-competitive initiatives. We and many others from the AI research and innovation fields would make immediate use of such data sets and would develop further innovations that are otherwise hard to assess.
At Genedata, our AI R&D is rooted in strong and productive industry collaborations and customer partnerships, which also provide quality data. A number of our customers partner with us and provide access to various high-quality, production-relevant data sets, which cover a wide range of bioassays and experimental conditions.
Matt Segall, Ph.D.: Yes, quality data is key to the successful application of AI. Easily accessible data in the public domain tends to be very noisy and not representative of data generated in ongoing drug discovery projects. We are tackling this in the following ways:
1. We are working collaboratively with large pharma and biotech to develop and validate our AI approaches using their proprietary data. In addition to providing proof of concept in practical applications, these projects also deliver immediate value to our collaborators.
2. We are working with Medicines Discovery Catapult (MDC), as part of the DeepADMET collaboration. MDC are applying ML and natural language processing to identify valuable, but difficult-to-find, data from the scientific literature to supplement those in existing, curated databases.
3. We are developing deployable platforms that will be integrated with our customers’ data sources (behind their firewalls) to apply our unique DL methods routinely to their proprietary data and make the results seamlessly available to researchers in their organisation. This will ensure that the resulting DL models are built on high-quality, relevant compounds and data for their discovery projects.
Biocompare: Are there any other challenges that are impacting broader usage of AI in drug discovery?
Grant Wishart, Ph.D.: Additional challenges within the industry, perhaps more pertinent within small to mid-sized biotech, can be distilled down to capital investment in AI technologies, whether or not to build internal capabilities, or to access it through partnerships with dedicated technology companies and/or CROs. Healthy skepticism will also be a hurdle in AI methodologies achieving broader proliferation, particularly within conservative organizations. This will gradually be alleviated through demonstrated success stories on live drug discovery programs and a more complete understanding of the applications and circumstances where the technology has the most impact and those which lead to failure.
Stephan Steigele, Ph.D.: While I’ve observed that AI-based algorithms in many drug discovery projects have outstanding performance, they also demand more testing before a broader, more generic application is possible. Currently, they lack a commonly accepted standard to stress-test and measure AI applications and so there either is blind trust in the procedure or no trust at all. From our own research, we learned how tough it is to generalize AI functionalities. In our domain, life science, research must cope with huge signal variability, which oftentimes is not covered by the available training data. This is a challenge—particularly when it comes to clinical decision-making that affects lives.
Nevertheless, AI is unstoppable and will fuel a growing range of applications in drug discovery. All the challenges I’ve mentioned will be solved in the coming years. As scientists better understand AI, standards will evolve, and more robust applications will come with an increasing acceptance of AI solutions as a routine part of drug discovery.
Matt Segall, Ph.D.: One challenge is the justifiable scepticism among researchers about whether AI methods will deliver on their promise. There is a lot of hype about AI methods and some early attempts have failed to produce the anticipated results in practice. Therefore, rigorous proof of concept is necessary for successful adoption of new methods.