Knowing which DNA sequences interact with which proteins can reveal a lot about chromatin organization and the role of histone post-translational modifications— and with it about enhancers and promoters, effects of mutations and interventions, transcriptional networks, the location of unannotated genes, and generally how expression is regulated. Perhaps the most enduring methods used to probe these interactions over the past three-plus decades are based on immunoprecipitation of DNA-interacting proteins with subsequent examination of the associated DNA. Here we look at the role of chromatin immunoprecipitation (ChIP) and how it relates to competing and complementary techniques to characterize the DNA sequences associated with genomic control.
What is ChIP?
In its simplest iteration, ChIP uses antibodies against DNA-binding proteins such as histones or transcription factors to capture the DNA associated with it. Cells are often first treated with a reversible cross-linking agent such as formaldehyde, with longer cross-linkers used to interrogate higher-order interactions (such as with proteins that associate only indirectly to DNA). And in native ChIP, tight interactions such as with histones can be interrogated without any fixation. Cells are then lysed, the chromatin is fragmented by sonication or nuclease treatment, protein-DNA complexes are captured, and the DNA is released.
DNA sequences are then interrogated by qPCR (ChIP-qPCR) or next-generation sequencing (ChIP-seq). For more focused studies in which the targets are known, qPCR may be the assay of choice. But “there is a cost breakpoint, when the number of binding sites you want to analyze starts to exceed 100, then it’s cheaper to do sequencing,” points out Tae Hoon Kim, associate professor of biological sciences at the University of Texas at Dallas.
ChIP-on-chip (also called ChIP-chip), in which DNA microarrays are used to assay the sequences pulled down by the immunoprecipitation, was another popular readout. “But as soon as high-throughput sequencing came to be in the mid-2000s that got displaced by ChIP-seq,” notes Trey Ideker, professor of medicine at the University of California San Diego.
Kim says that there is a standard algorithm, Model-based Analysis for ChIP-seq (MACS), that allows users to call the binding sites from the sequencing data. And while the resolution of ChIP is limited in part by fragment size—in the hundreds of base pairs—“there are some computational tools that basically pinpoint the actual center of the binding site” based on the asymmetry of reads recovered from the top versus the bottom strands.
“A much quicker road to high-resolution binding information is if you simply [use an exonuclease to] chew away all parts of the DNA that aren’t stuck to the factor,” Ideker says, explaining the technique called ChIP-exo. His lab now routinely uses ChIP-exo any time they perform ChIP—it’s “a simple trick that works really well.”
It’s the antibody, stupid
The DNA sequences pulled down by ChIP are dependent on the antibodies used to recognize the associated proteins. These should be validated to work in ChIP, with verified specificity. It’s important to optimize stringency of the precipitations, running both positive and negative controls to assure that, for example, the desired transcription factor or histone mark is being targeted.
If a good antibody is not available, in many cases a fusion of an epitope tag with the protein of interest (POI) can be genetically engineered, allowing an antibody against the tag to be used in its stead. In the case of CETCh-seq, CRISPR/Cas9 is used to engineer the tag onto its target. And Active Motif’s enChIP® uses a deactivated CRISPR/Cas9 system “to bind a very targeted, unique site in the human genome and pull that down to see where that site might be interacting with other genomic loci, or to see if there are background target sites for CRISPR/Cas9 guide RNA,” says Adam Blattler, who develops ChIP kits and other genomics assays for the company.
 There are a host of other related and hyphenated techniques used to probe DNA-protein interactions. For example, in ChIP-re-ChIP, immunoprecipitation is sequentially performed using antibodies to different proteins in order to demonstrate that the proteins associate with the same region of DNA (although the authors caution that this does not prove that the proteins associate with each other).
There are a host of other related and hyphenated techniques used to probe DNA-protein interactions. For example, in ChIP-re-ChIP, immunoprecipitation is sequentially performed using antibodies to different proteins in order to demonstrate that the proteins associate with the same region of DNA (although the authors caution that this does not prove that the proteins associate with each other).
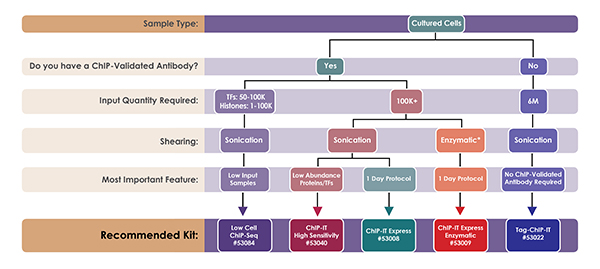
Image: The features and limitations of ChIP kits. Image courtesy of Active Motif.
With CUT&RUN, meanwhile, a nuclease/Protein A fusion attaches to antibodies bound to the POI. “Essentially you make cuts surrounding those binding sites, releasing DNA,” Blattler explains. “It gives the same answer as ChIP, but it’s not an immunoprecipitation.” CUT&RUN has, in some cases, been found to be more sensitive, and involves fewer steps, than ChIP, and “is still being evaluated by many in the field for widespread application akin to ChIP.”
A fair number of researchers follow their own or established protocols to perform ChIP experiments. In addition, many kits exist, or are being developed, to perform ChIP, for example, on low cell numbers, on FFPE samples, in high throughput, or even to add sequencing library adapters during enrichment (TAM-ChIP™). Services exist that will do some, or all, of it for you as well.
Chromatin architecture
Danette Daniels, Promega’s group leader in functional proteomics, has noticed that ChIP’s importance at epigenetics meetings “is not as heavily weighted as it used to be,” with more emphasis being placed on chromatin accessibility assays (such as ATAC-seq) as well as conformation assays.
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) takes advantage of the ability of transposons to randomly integrate into open chromatin, suggesting where transcription factors can bind and DNA regulation may be taking place. And there are a host of chromosome conformation capture (3C) assays that use crosslinking, enzymatic digestion, and intramolecular ligation to interrogate relations between non-linearly connected stretches of chromatin such as enhancer-promoter interactions.
ChIP “identifies a ton of different DNA elements that are bound by your transcription factor of interest and a lot of those sites are probably not functional,” opines Ideker. To study transcription networks, he adds approaches like CRISPR knockdowns and expression quantitative trait loci (eQTL) mapping. “You can say that they complement each other. One is physical, the others are functional. Together, the body of evidence starts to look solid.”